RNA Aptamer Targeting to Protein with Single Amino Acid Mutation Screened out
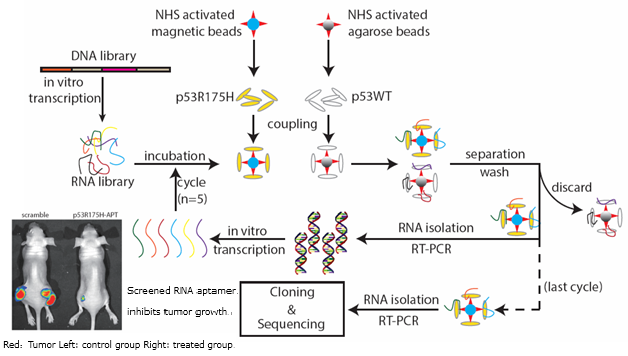
On July 27th, PNAS announced a new screening method to screen out nucleic acid (RNA) aptamer targeting protein with subtle changes (e.g. single amino acid residues mutation). The research came from Prof. SHAN Ge’s team from University of Science and Technology of China. The paper is entitled ‘The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation’.RNA aptamer is an artificially synthesized non-coding RNA through a screening strategy called SELEX. It can specifically bind to small molecules, proteins and even whole cells. The screening of RNA aptamer utilizes a sufficiently diverse RNA library, which harbors potential of binding diversity and specificity just like antibody. The concept and technique of RNA aptamer is first introduced by Jack Szostak (2009 Nobel laureate in Physiology&Medicine) in 1990. Yet there was report about an RNA aptamer specific to a protein with a single amino acid changeover a decade.
Taking a p53 single mutation protein, which is frequently found in tumors, as a target, Prof. Ge Shan and his team used a previously unidentified screening method, and screened out an RNA aptamer. This RNA aptamer specifically recognizes mutant p53 protein but not its wild-type. What’s more meaningful, this RNA aptamer can effectively inhibit the pro-tumor function of mutant p53 protein in both cultured cells and nude mice bearing tumors.
The first author of this publication is Dr. Liang Chen, and this paper is the major part of his PhD work. This study received help from Prof. Jun Wang and Prof. Tao Zhu’s lab, School of Life Sciences, USTC. This work is funded by Ministry of Science and Technology, Chinese Academy of Sciences, National Natural Science Foundation of China and USTC Non-coding RNA Function and Mechanism Innovation Group. Recent years in non-coding RNA research field, Prof. Ge Shan’s lab has published several significant research articles in prestigious journals such as Nature Communications (2012), Nature Structural & Molecular Biology (2015), etc.
Link to this paper:http://www.pnas.org/content/early/2015/07/22/1502159112.abstract
To whom correspondence should be addressed. Email: shange@ustc.edu.cn.
(HUANG Jun, School of Life Science, Department of Scientific Research)
Back
