USTC Reveals Epigenetic Regulatory Mechanism on Cutaneous T cell Lymphoma
Gene regulation is a complex system of genetic and epigenetic modifications, and in-depth studies of patients' epigenome have very important scientific implications for understanding the pathogenesis of many diseases at the molecular level, optimizing diagnosis and treatment programs. This article is the first real-time construction of personalized epigenetic regulatory networks in tumor cells in vivo. On the one hand, this work established a template for mechanistic studies on personalized epigamic regulation in other diseases, and on the other hand, provided scientific basis for precise medical suggestions for patients with CTCL. Thus has a very important scientific significance to promote the development of precision medicine.
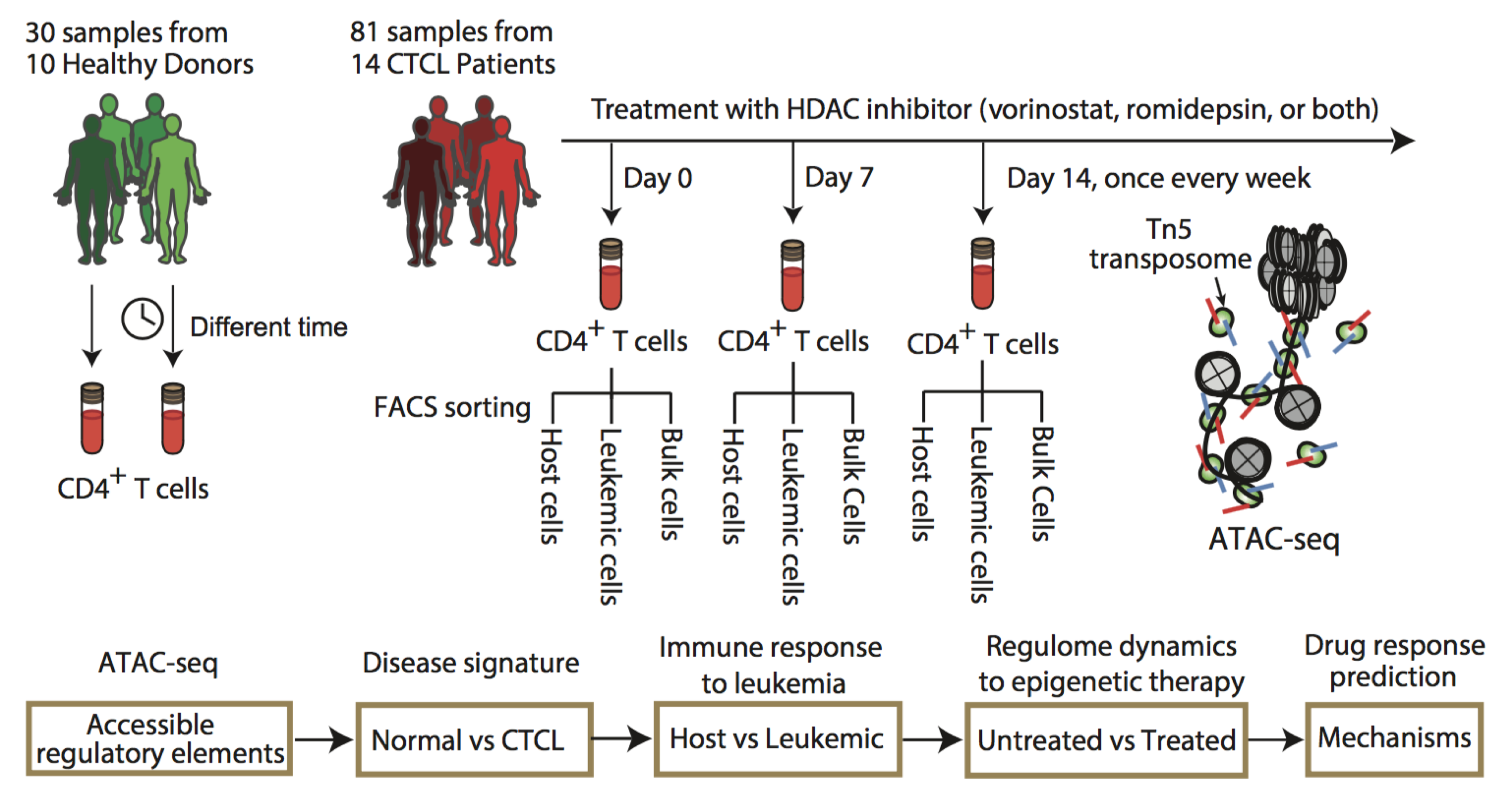
In this study, the authors used a new technique called ATAC-seq to study the mechanism of individualized epigenetic regulation of CTCL and its decisive effect on drug sensitivity in human T-cell lymphoma (CTCL). CTCL is a rare and complex disease caused by clonal hyperplasia of T lymphocytes. Late stages of CTCL, such as Sézary syndrome, cancer cells can be transferred to the bloodstream, leading to T cell leukemia, and makes it a very fatal disease. In this paper, the authors isolated the CD4+ T cells from normal and CTCL patients’ fresh blood using Fluorescence-activated cell sorting (FACS). Then applying the ATAC-seq technique to rapidly detect the open chromatin sites from the normal and leukemic T lymphocytes in vivo. By integrating a variety of genomic information and bioinformatics approaches, the authors in-depth analyzed the landscape epigenetic signatures of CTCL, and constructed tumor, patient and cell type specific gene regulatory networks. The authors also studied the dynamics of the patients’ epigenomes during histone deacetylase inhibitor (HDACi) anticancer drugs treatment at a weekly resolution, and revealed the dynamic regulatory mechanisms of epigenome and key transcription factors on the patient's drug sensitivity, and successfully constructed models to accurately predict the patients’ drug sensitivity on HDACi anti-cancer drugs.
Figure. Schematic outline of the study design shows 30 samples from 10 healthy donors and 81 samples from 14 CTCL patients under HDACi therapy (top), with a bioinformatics pipeline for data analysis (bottom). Host and leukemic cells were isolated from FACS based on CD3+CD4+Vβ− and CD3+CD4+Vβ+, respectively. ATAC-seq was then performed on normal CD4+, CTCL, leukemic, and host cells. (Image by QU Kun)
This study was published online at Cancer Cell, one of the internationally renowned scientific journals in cancer research, entitled “Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors”. Prof. Qu at the school of life science is the first author, and USTC is the first institute. Undergraduate student JIN Yonghao and lab research assistant JIN Chen also contributed in this study. This work is supported by research funding from the Organization Department of the CPC Central Committee, the Department of Science and Technology, the National Science Foundation of China, the Chinese Academy of Science and the USTC startup.
(School of Life Science and Medical Center)
The link of the paper:
http://www.cell.com/cancer-cell/fulltext/S1535-6108(17)30204-0
Contact
Prof. QU Kun
Back