Researchers Reveal Structural Mechanism of Tetrahymena Ribozyme Self-Splicing Reaction
A team led by Prof. ZHANG Kaiming from the University of Science and Technology (USTC) solved six conformations in the second-step self-splicing of Tetrahymena ribozyme using cryogenic electron microscopy (cryo-EM), revealing the mechanism of group 1 introns coordinating self-splicing reactions. Their work was published in Nature Communications on March 16 th .
GroupⅠintrons are catalytic RNAs that can fold into tertiary structure with a active site with the help of metal ions, thereby promoting catalysis without proteins. However, the analysis of RNA structures faces enormous challenge due to the heterogeneity and flexibility of RNA. Since the discovery of Tetrahymena group I introns, numerous studies have elucidated its underlying catalytic mechanism, but the 3D structure of the full-length molecule and its rearrangements are yet to be obtained.
To resolve the structural mechanism and the conformation changes in the self-splicing reactions, Prof. ZHANG Kaiming’s team first designed the substrates for the self-splicing reactions. After verifying that the substrates could complete the catalytic reactions and determining the optimal reaction conditions, the team captured structure snapshots of different conformations of Tetrahymena ribozyme during the self-spicing process using single-particle cryo-EM analysis.
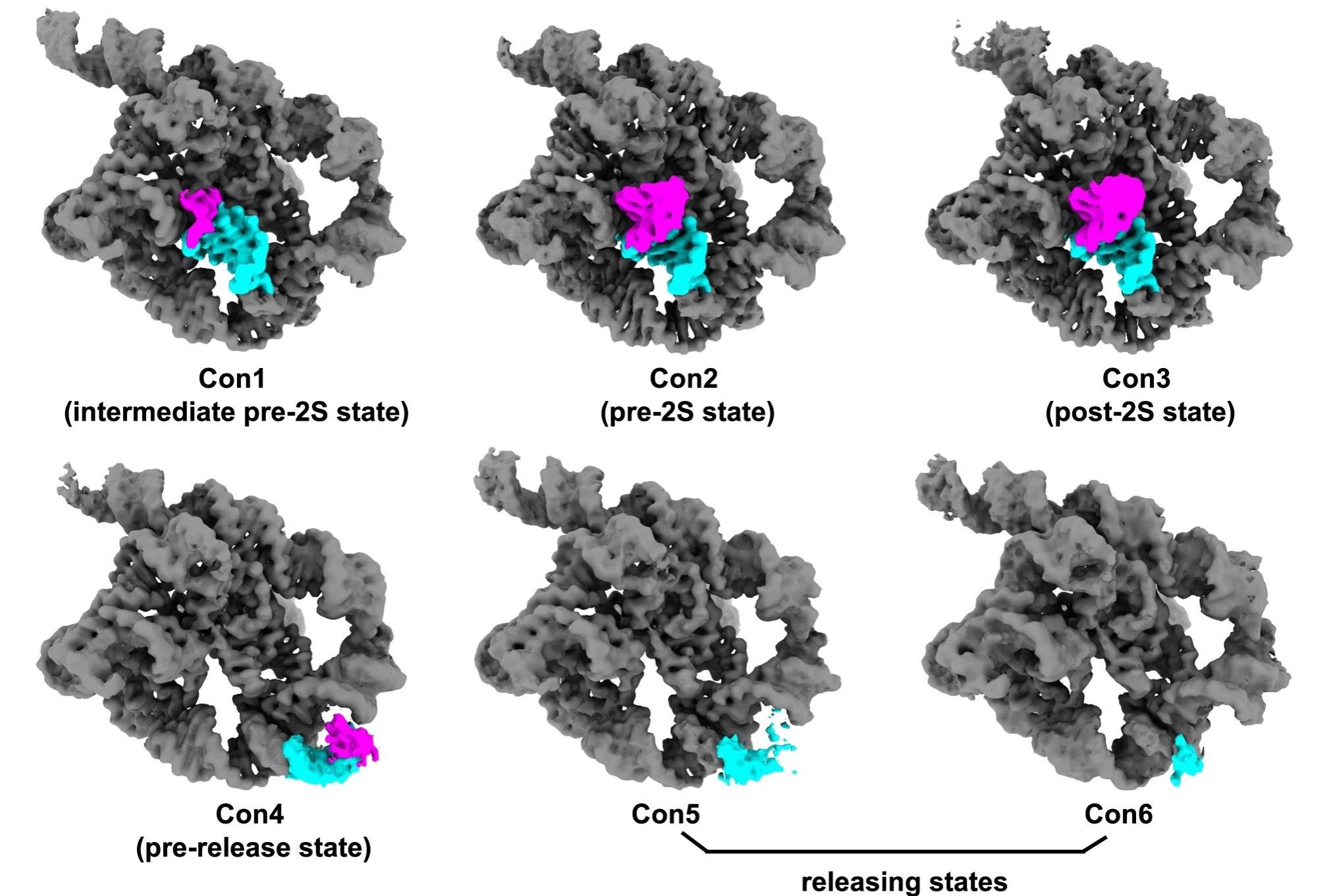 Different conformations of Tetrahymena ribozyme in the self-splicing reactions. (Image by LI Shanshan et al.)
Different conformations of Tetrahymena ribozyme in the self-splicing reactions. (Image by LI Shanshan et al.)
The first step starts from oligonucleotide substrate base pairing with the apoenzyme to form the P1-P1 extension duplex. After that, the tertiary interactions between P1 and three single-stranded segments within the catalytic core mediate docking of the duplex into the active site. Then the oligonucleotide product of the first step is released. The second step is the ligation reaction, where the 3’-exon substrates become docked in the ribozyme through tertiary contacts and metal coordination. Then the reaction goes gradually to an intermediate period, where metal coordination weakens and the tertiary interactions are lost. The exons are ligated in this period. Sequentially, the ribozyme relaxed and the product duplex undocks, reaching the substrate-free site. Finally, the duplex is unpaired and the product is released. Furthermore, the researchers also found structural evidences for the rearrangements of some hydrogen bonds and metal ions on active sites, which promote catalysis and coordinate self-splicing reactions.
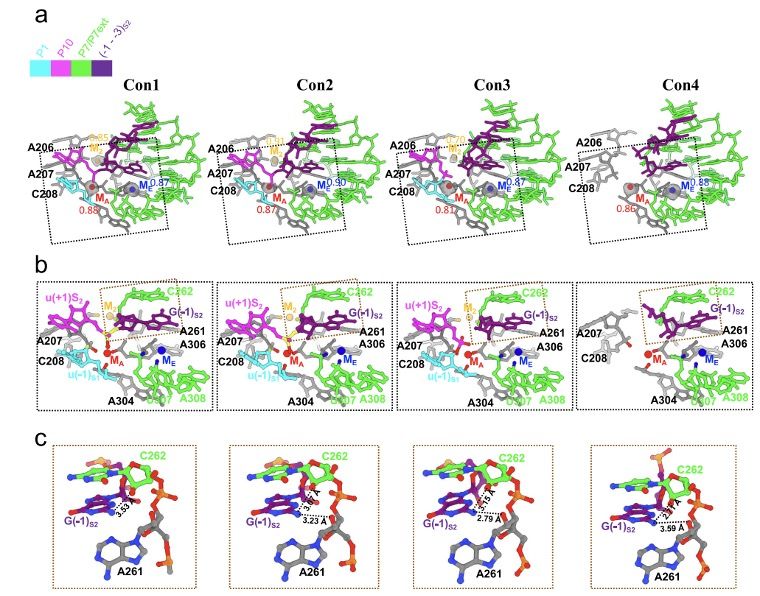 Comparison of active sites among different splicing states. (Image by LI Shanshan et al.)
Comparison of active sites among different splicing states. (Image by LI Shanshan et al.)
This research revealed the mechanism of self-splicing reaction on atomic level, proving the advantages of using cryo-EM to resolve RNA structure. With the increasing application of cryo-EM in the research of structurally heterogeneous molecules, it will become a vital tool in many other biological research like the folding of RNA systems.
Paper link: https://www.nature.com/articles/s41467-023-36724-5
(Written by WENG Guchcen, edited by ZHANG Yihang, USTC News Center)
Back
